truQuant
The truQuant tool is used to build an annotation of transcribed genes from PRO-Seq data. It also quantifies the
pause regions and gene bodies from the generated annotation while blacklisting enhancers, downstream promoters, and
non RNA Polymerase II transcripts.
Note
This tool requires bedtools be installed.
Usage and option summary
Usage:
PolTools truQuant [-h] [-a [annotation_extension]]
[-b [blacklisting_percent]]
[-r [pause_region_radius]] [-t [threads]]
[-d [min_seq_depth]]
[-m [min_avg_transcript_length]]
[-l [max_fragment_length]]
sequencing_file_for_annotation
[sequencing_files [sequencing_files ...]]
Required Arguments |
Description |
|---|---|
Sequencing file for annotation |
Bed formatted file from a sequencing experiment. |
Optional Arguments |
Description |
|---|---|
-a, –annotation_extension |
Distance of base pairs to extend the 5’ end of all genes upstream. Default is 1000. |
-b, –blacklisting_percent |
Percentage (number between 0 and 1) of reads in the pause region that is necessary to blacklist a TSR in the gene body. For example, a pause region with 100 reads and a blacklist percentage of 0.3 means a TSR in the gene body needs at least 30 reads to be blacklisted. Default is 0.3. |
-r, –pause_region_radius |
Base pair amount to go upstream and downstream centered on the avgTSS. The pause region will be of size 2 * pause region radius. Default is 75. |
-t, –threads |
Maximum number of threads to run truQuant. Default is the max available on the system. Please note that truQuant will use a maximum of 46 threads for finding TSRs and a one thread for each sequencing file (these two processes do not happen at the same time). |
Sequencing Files |
Additional sequencing files can be provided to be quantified using the generated annotation. The files will be blacklisted then quantified using the number of 5’ end reads in the pause region and the number of 3’ end reads in the gene body. |
-d, –min_seq_depth |
The minimum number of 5’ reads to be considered as a TSR in tsrFinder |
-m, –min_avg_transcript_length |
The minimum average transcript length will eliminate TSRs from sequencing artifacts in tsrFinder |
-l, –max_fragment_length |
The maximum transcript length for a read to be included in tsrFinder |
Behavior
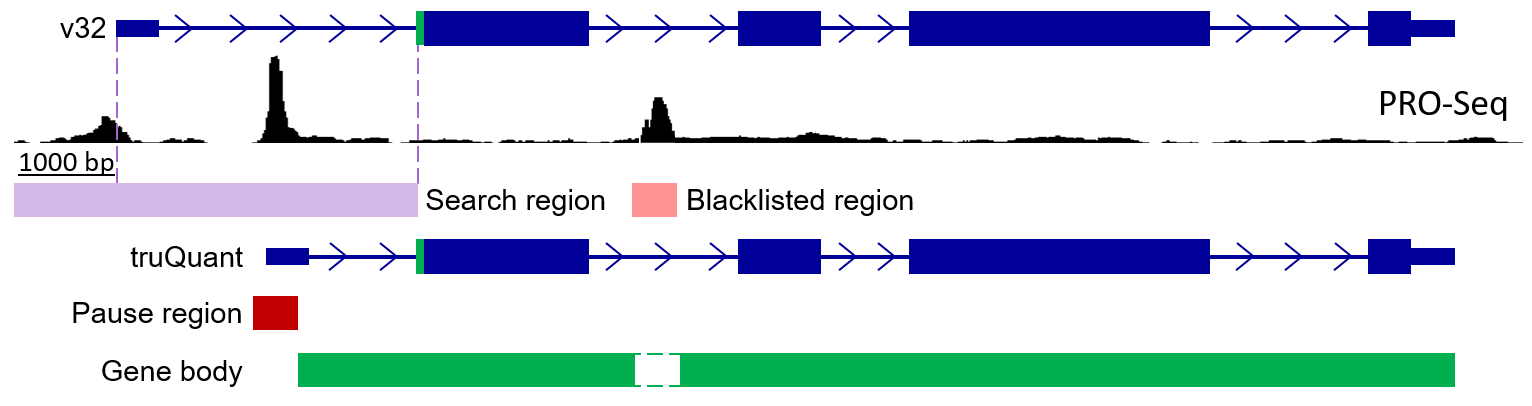
truQuant will generate search regions 1000 bp upstream of the 5’ end of protein coding genes from GENCODE v32. Then,
tsrFinder will be run to determine the max TSR in the search region. Inside this TSR, the max TSS will be chosen as the
annotated 5’ end. The pause region will be the 150 bp region surrounding the weighted average TSS (avgTSS) and gene body
will be the end of the pause region to the TES. TSRs in the gene with more than 30% of the reads as the max TSR will be
blacklisted. 5’ ends in the pause regions will be quantified and 3’ ends in the gene bodies will be quantified.
For example:
$ head -n 5 seq_file.bed
chr1 11981 12023 A00876:119:HW5F5DRXX:1:2168:2248:1407 255 -
chr1 13099 13117 A00876:119:HW5F5DRXX:1:2203:31403:26757 255 -
chr1 13356 13423 A00876:119:HW5F5DRXX:1:2151:15808:7827 255 -
chr1 13435 13477 A00876:119:HW5F5DRXX:1:2273:15781:19241 255 -
chr1 13739 13772 A00876:119:HW5F5DRXX:1:2256:29966:10520 255 -
$ PolTools truQuant seq_file.bed
$ head -n 1 control-truQuant_output.txt
Gene Chromosome Pause Region Left Pause Region Right Strand Total 5' Reads MaxTSS MaxTSS 5' Reads Weighted Pause Region Center STDEV of TSSs Gene Body Left Gene Body Right Gene Body Distance seq_file.bed Pause Region seq_file.bed Gene Body
NOC2L chr1 959177 959327 - 194 959255 46 959250 13.306459171023036 944203 959177 14974 194 18
KLHL17 chr1 960552 960702 + 234 960632 27 960626 25.417791063821863 960702 965719 5017 234 17
PLEKHN1 chr1 966439 966589 + 25 966521 8 966513 19.47408534437497 966589 975865 9276 25 11
HES4 chr1 1000013 1000163 - 239 1000096 87 1000086 27.14758979723915 998962 1000013 1051 239 68
ISG15 chr1 1000204 1000354 + 160 1000295 12 1000278 36.24344768368484 1000354 1014540 14186 160 111
AGRN chr1 1020042 1020192 + 112 1020119 35 1020116 25.189637892253575 1020192 1056118 35926 112 76
RNF223 chr1 1074208 1074358 - 32 1074306 10 1074284 32.567238138964136 1070967 1074208 3241 32 8